DeconSeq. DECONtamination of SEQuence data.
Manual
Web versionNecessary resources
Upload data
Standalone version
Necessary resources
Available options
Sequence contaminant removal
How to create a database

Preprocessing 454 data
01/16/2011
PDF, 92 KB

Preprocessing FASTA-only data
01/16/2011
PDF, 76 KB

Raw data processing
01/16/2011
PDF, 112 KB

Quality control
01/16/2011
PDF, 520 KB

Data preprocessing
09/01/2011
PDF, 827 KB
Introduction
Sequences obtained from impure nucleic acid preparations may contain DNA from sources other than the sample. Those sequence contaminations are a serious concern to the quality of the data used for downstream analysis, possibly causing misassembly of sequence contigs and erroneous conclusions. Therefore, the removal of sequence contaminants presents a necessary step for all metagenomic projects.
DeconSeq is distributed under the GNU Public License (GPL). All its source codes are freely available to both academic and commercial users. The latest version can be downloaded at the SourceForge download page.
Web version
TOP OF PAGEThe interactive web interface of DeconSeq can be used to automatically detect and efficiently remove sequence condaminations from genomic and metagenomic datasets.
Necessary resources
Hardware
- Computer connected to the Internet
Software
- Up-to-date Web browser (Firefox, Safari, Chrome, Internet Explorer, ...)
Files
- FASTA file with sequence data
- FASTQ file (as alternative format to trim sequence and quality data)
Upload data to the DeconSeq web version
To upload a new dataset in FASTA or FASTQ format to DeconSeq, follow these steps:
1. Go to http://deconseq.sourceforge.net
2. Click on "Use DeconSeq" in the top menu on the right (the latest DeconSeq web version should load)
3. Select your FASTA or FASTQ file
4. Select the retain and remove (optional) database(s)
5. Click "Submit"
Notes
Step 4 allows selecting two types of reference databases. The remove databases are the databases used to screen for contaminants. The retain databases are used to eliminate redundant hits with higher similarity to non-contaminant genomes (e.g. viral sequences in the human genome for viral metagenomes).
Sequence contaminant removal
TOP OF PAGESequences obtained from impure nucleic acid preparations may contain DNA from sources other than the sample. Those sequence contaminations are a serious concern to the quality of the data used for downstream analysis, possibly causing erroneous conclusions.
The dinucleotide abundance approach used by PRINSEQ to identify if a dataset contains contamination does not allow the identification and removal of single contaminant sequences. Sequence similarity seems to be the only reliable option to identify single contaminant sequences. However, BLAST searches against the human reference genome are slow and lack corresponding regions (gaps, variants, ...). Furthermore, novel sequences were found in every new human genome sequenced [1]. DeconSeq allows the automated identification and removal of sequence contamination in longer-read datasets (>150 bp mean read length) using an algorithm tens of times faster than BLAST [2].
DeconSeq uses the query sequence coverage and alignment identity to identify sequences that are similar to a contaminant sequence in the remove databases. The identity is a measure for how similar the query sequence to the reference sequence is and the coverage is a measure of how much of the query sequence is similar to the reference sequence. If a retain database is selected, query sequences are classified as "Similar to Both" if they are similar to sequences in the remove and retain database using the coverage and identity thresholds. All other query sequences can be classified as "Clean" or "Contamination", as shown below.

DeconSeq generate Coverage vs. Identity plots to guide users in their selection of threshold values. The plots show the number of matching reads for different query coverage and alignment identity values. The number of matching reads with a specific coverage and identity value defines the size of each dot in the plots. Red dots represent matching reads against the remove databases and blue dots against retain databases. The column and row sums at the top and right of each plot allow an easier identification of the number of sequences that match for a particular threshold value.
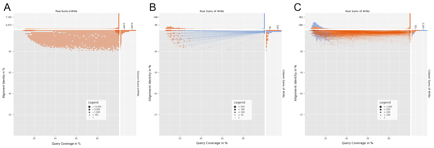
The plots for matching reads against the remove databases do not show matching reads that additionally have a match against the retain databases (A). Results for reads matching against both databases are shown in a second plot where dots for a single read are connected by lines. If the match against the remove database is more similar, then the line is colored red, otherwise blue. In B, for example, the majority of sequences is more similar to the retain databases and in C the majority is more similar to the remove databases.
References
1. Li R, Li Y, Zheng H, Luo R, Zhu H, Li Q, Qian W, Ren Y, Tian G, Li J, Zhou G, Zhu X, Wu H, Qin J, Jin X, Li D, Cao H, Hu X, Blanche H, Cann H, Zhang X, Li S, Bolund L, Kristiansen K, Yang H, Wang J, Wang J: Building the sequence map of the human pan-genome. Nat. Biotechnol 2010, 28:57-63.
2. Li H, Durbin R: Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26:589-595.
Standalone version
TOP OF PAGEThe standalone version does not provide any graphical outputs and databases. The databases used for contamination screening have to be generated as described in the readme file distributed with the source code. The readme file also contains information on the usage of the standalone version.
Necessary resources
Hardware
- Computer with a Linux/Unix or Mac OS X operating system
Software
- DeconSeq standalone (available at http://deconseq.sourceforge.net)
- Perl 5 (or higher)
Files
- FASTA file with sequence data
- FASTQ file (as alternative format)
Available options
|
Option/flag |
Description |
Default |
Range |
-help or -h |
Print the help message; ignore other arguments |
||
-man |
Print the full documentation; ignore other arguments |
||
-version |
Print program version; ignore other arguments |
||
| Input/Output options | |||
-show_dbs |
Prints a list of available databases |
|
|
-f |
Input file in FASTA or FASTQ format that contains the query sequences |
|
FILE |
-dbs |
Name of remove database(s) to use. Names are according to their definition in the config file. Separate multiple database names by comma without spaces. |
hsref |
STRING |
-dbs_retain |
Output format |
|
STRING |
-out_dir |
Directory where the results should be written. If the directory does not exist, it will be created. |
Current directory | STRING |
-group |
If dbs_retain is set, then this option can be used to group the sequences similar to -dbs and -dbs_retain databases with either the clean or the contamination output file. If -group is not set and -dbs_retain is set, then three separate files will be generated. |
[1,2] |
|
-no_seq_out |
Prevents the generation of the FASTA/FASTQ output file for the given coverage and identity thresholds. This feature is e.g. useful for the web-version since -i and -c are set interactively and not yet defined at the data processing step. |
|
|
-keep_tmp_files |
Prevents from unlinking the generated tmp files. These usually include the id file and the .tsv file(s). This feature is e.g. useful for the web-version since .tsv files are used to dynamically generate the output files. |
|
|
-id |
Optional parameter. If not set, ID will be automatically generated to prevent from overwriting previous results. This option is useful if integrated into other tools and the output filenames need to be known. |
|
STRING |
| Alignment options | |||
-i |
Alignment identity threshold in percentage. The identity is calculated for the part of the query sequence that is aligned to a reference sequence. For example, a query sequence of 100 bp that aligns to a reference sequence over the first 50 bp with 40 matching positions has an identity value of 80%. |
|
INT [1..100] |
-c |
Alignment coverage threshold in percent. The coverage is calculated for the part of the query sequence that is aligned to a reference sequence. For example, a query sequence of 100 bp that aligns to a reference sequence over the first 50 bp with 40 matching positions has an coverage value of 50%. |
|
INT [1..100] |
-S |
Chunk size of reads in bp as used by BWA-SW |
10000000 |
INT |
-z |
Z-best value as used by BWA-SW |
1 |
INT |
-T |
Alignment score threshold as used by BWA-SW |
30 |
INT |
How to create a database
The following example shows how to create a database from the human reference genome for the use with BWA, BWA-SW and DeconSeq. The commands used below assume a Unix-based operating system.
Download sequence data
There are several places where one can retrieve the sequence data. The human reference genome build 37 can be downloaded from the National Center for Biotechnology Information (NCBI) FTP server. If you get an error, check if the files are still available at the specified loacation and if the file names are still the same.
$ for i in {1..22} X Y MT; do wget ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/Assembled_chromosomes/seq/hs_ref_GRCh37.p2_chr$i.fa.gz; doneExtracting and joining data
The sequence data is compressed with the GZIP algorithm and in 25 different files. The next command will extract the data and write them to a single file. At the same time, the compressed files will be deleted (rm command). The -v in the gzip command provides an easy option to see the progress.
$ for i in {1..22} X Y MT; do gzip -dvc hs_ref_GRCh37.p2_chr$i.fa.gz >>hs_ref_GRCh37_p2.fa; rm hs_ref_GRCh37.p2_chr$i.fa.gz; doneSplitting sequences by long repeats of ambiguous base N
BWA and BWA-SW replace ambiguous bases (everything that is not an ACGT) with a random base (an ACGT). This can, for example, result in false positive hits in long stretches of Ns that were randomly converted into a sequence similar to a query sequence. The reason for the replacement of ambiguous bases is the 2 bit representation of sequence data that only allows to store/represent four different bases (as 00, 01, 10 and 11). Therefore, it is a good idea to remove long strechtes of ambiguous bases before creating the database (especially considering the stretches of 50,000 Ns in the human reference genome). The following command will additionally remove Ns at the beginning and end of each sequence (e.g. the 10,000 Ns at the beginning of chromosome 1).
$ cat hs_ref_GRCh37_p2.fa | perl -p -e 's/N\n/N/' | perl -p -e 's/^N+//;s/N+$//;s/N{200,}/\n>split\n/' >hs_ref_GRCh37_p2_split.fa; rm hs_ref_GRCh37_p2.faFiltering sequences
BWA-SW uses the concatenation of all input sequences when generating the database. After splitting the genomic sequences, it is possible that a query sequence will be longer than a sequence in the database. This can cause problems when a query sequence aligns over multiple (concatenated) sequences in the database. In the current version, BWA-SW is not able to resolve alignments spanning more than two database sequences. By removing shorter sequences, a query sequence should not be able to align to more than two sequences in the database.
In addition to short sequences, sequence duplicates should be removed as well. Duplicates do not add new information, but add to the size of the database. In order to reduce the amount of false positive alignments (see above), sequences with too many ambiguous bases should be removed.
The sequences can be easily filtered with programs such as PRINSEQ (http://prinseq.sourceforge.net). The following command will additionally rename the sequence identifiers to ensure unique identifiers in the whole data set and delete the input file (rm command).
$ perl prinseq-lite.pl -log -verbose -fasta hs_ref_GRCh37_p2_split.fa -min_len 200 -ns_max_p 10 -derep 12345 -out_good hs_ref_GRCh37_p2_split_prinseq -seq_id hs_ref_GRCh37_p2_ -rm_header -out_bad null; rm hs_ref_GRCh37_p2_split.faCreating the database
The BWA program provides an option to create databases from sequence data in FASTA format. For bigger data sets, the bwtsw algorithm should be used. The maximum size of a database is 4GB. If you want to generate a database from larger data sets, you have to split the data into chunks of less than 4GB. The indexing process can take a while and therefore, we will use the "&" at the end of the command to run the process in the background. In order to know what BWA is doing, we will write its output to the file bwa.log. You can use "$ tail -f bwa.log" to check for the current status.
$ bwa index -p hs_ref_GRCh37_p2 -a bwtsw hs_ref_GRCh37_p2_split_prinseq.fasta >bwa.log 2>&1 &When the BWA program has finished, you should have eight files with the name specified by the -p parameter.
